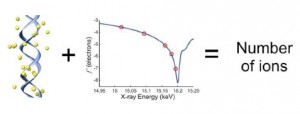
Counting ions around DNA with ASAXS
S. A. Pabit, S. P. Meisburger, L. Li, J. M. Blose, C. D. Jones, L. Pollack
J. Am. Chem. Socl, 132 (46), pp 16336 (2010).
ABSTRACT
The majority of charge compensating ions around nucleic acids forms a diffuse counterion “cloud” that is not amenable to investigation by traditional methods that rely on rigid structural interactions. Although various techniques have been employed to characterize the ion atmosphere around nucleic acids, only Anomalous Small- Angle X-ray Scattering (ASAXS) provides information about spatial distribution of the ions. Here, we present an experimentally straightforward extension of ASAXS to count the number of ions around nucleic acids.
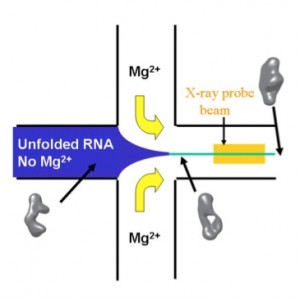
Time resolved SAXS and RNA folding
L. Pollack
Biopolymers 95, 543 (2011).
ABSTRACT
Small angle x-ray scattering provides low resolution structural information about macromolecules in solution. When coupled with rapid mixing methods, SAXS monitors time-dependent conformational changes of RNA induced by the addition of Mg2+ to trigger folding. This time-resolved SAXS provides unique information about the global or overall structures of transient intermediates populated during folding. When couple with results from methods that provide local measures of folding, a more complete picture of the process emerges. Together, these methods highlight the interplay of electrostatic repulsion, which biases the molecules towards more extended states, and tertiary contact formation which favors more compact states.