Lab on a Chip, 2014: Microfluidic immunocapture of circulating pancreatic cells using parallel EpCAM and MUC1 capture: characterization, optimization and downstream analysis
Citation: Thege FI, Lannin TB, Saha TN, Tsai S, Kochman ML, Hollingsworth MA, Rhim AD and Kirby BJ. Kirby Microfluidic immunocapture of circulating pancreatic cells using parallel EpCAM and MUC1 capture: characterization, optimization and downstream analysis. Lab Chip (2014)doi pdf
Abstract: We have developed and optimized a microfluidic device platform for the capture and analysis of circulating pancreatic cells (CPCs) and pancreatic circulating tumor cells (CTCs). Our platform uses parallel anti-EpCAM and cancer-specific mucin 1 (MUC1) immunocapture in a silicon microdevice. Using a combination of anti-EpCAM and anti-MUC1 capture in a single device, we are able to achieve efficient capture while extending immunocapture beyond single marker recognition. We also have detected a known oncogenic KRAS mutation in cells spiked in whole blood using immunocapture, RNA extraction, RT-PCR and Sanger sequencing. To allow for downstream single-cell genetic analysis, intact nuclei were released from captured cells by using targeted membrane lysis. We have developed a staining protocol for clinical samples, including standard CTC markers; DAPI, cytokeratin (CK) and CD45, and a novel marker of carcinogenesis in CPCs, mucin 4 (MUC4). We have also demonstrated a semi-automated approach to image analysis and CPC identification, suitable for clinical hypothesis generation. Initial results from immunocapture of a clinical pancreatic cancer patient sample show that parallel capture may capture more of the heterogeneity of the CPC population. With this platform, we aim to develop a diagnostic biomarker for early pancreatic carcinogenesis and patient risk stratification.
Figures:
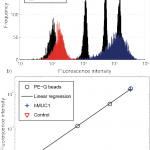
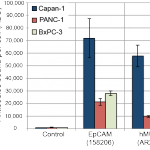
- Antibody bound per cell (ABC) counts for EpCAM, hMUC1 and PE labelled secondary control antibodies in cell lines Capan-1, PANC-1 and BxPC-3
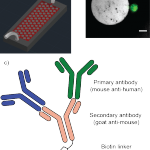
- a) Schematic overview of the GEDI device, modified from Kirby et al.,18 microposts not to scale b) calcein-stained (green) PANC-1 cell captured on a GEDI micro post functionalized with a cancer-specific MUC1 antibody. Scale bar: 20 μm c) silicon surface functionalization with primary antibodies and secondary antibody linker chemistry that allows for multiple parallel capture antibodies
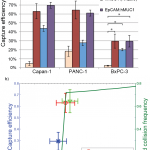
- a) GEDI microdevice immunocapture of Capan-1, PANC-1 and BxPC-3 cells by anti-EpCAM and anti-hMUC1 antibodies, and an anti- EpCAM/hMUC1 antibody cocktail, b) anti-EpCAM capture efficiencies of Capan-1, PANC-1 and BxPC-3 cells and the predicted collision frequency as function of cell size, based on previously described simulations.12 * indicates statistically significant difference with p = 0.05
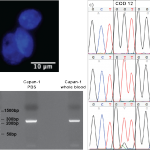
- a) Released and isolated DAPI-stained (blue) Capan-1 nuclei, b) agarose gel with a 249 bp Capan-1 KRAS RT-PCR product from cells spiked in PBS buffer and whole blood, c) Sanger sequencing of the KRAS codon 12 RT-PCR product from whole blood, Capan-1 cells spiked in PBS and Capan-1 cells spiked in whole blood. The whole blood sequence shows the wild type codon 12 genotype (GGT) while RNA extracted from Capan-1 cells spiked in PBS and whole blood shows the expected GGT → GTT oncogenic SNP. Note the presence of a small wild-type peak in the spiked blood sample. The presented sense sequences have been converted from anti-sense sequences generated by the Sanger sequencing reaction.
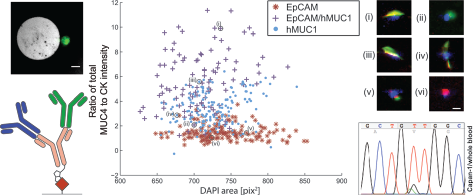
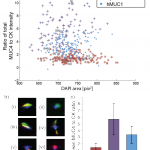
- Analysis of CPCs from a clinical PDAC patient sample, processed using anti-EpCAM, anti-EpCAM/hMUC1 and anti-hMUC1 GEDI devices, a) ratio of total MUC4 and CK staining intensity versus nuclear DAPI area in events classified as likely CPCs, b) inset images showing representative CPC events, i–ii anti-EpCAM/hMUC1 cocktail capture, iii–iv anti-hMUC1 capture and v–vi anti-EpCAM capture, images i, iii and iv are consistent with cells spread on microposts, DAPI (blue), cytokeratin (red), MUC4(green). Scale bar: 10 μm c) mean MUC4-to-CK total intensity ratios for all three capture modes, error bars indicate standard deviation within the population of identified CPCs.