Functional genomic diversification within single estuaries
One mechanism by which marine species with larval dispersal cope with highly heterogeneous and unpredictable environments is through high fecundity and sexually generated zygotic diversity. George Williams (1975) described this life history strategy as the ‘elm/oyster model’, emphasizing that meiotic recombination generates abundant phenotypic diversity among zygotes within each cohort. It follows that, in high fecundity organisms, physical stress gradients acting during dispersal and after settlement can generate large within-generation changes in genotype frequencies based on relative fitness. How much functional diversification results from within-generation selection within a single estuary, and to what extent does this dynamic maintain balanced polymorphisms?
Two recent projects in the Hare Lab investigated this question using genomic approaches. Laura Eierman is a recent PhD graduate who studied the functional genetics of osmoregulatory tolerances in the euryhaline oyster, Crassostrea virginica. Euryhaline describes the evolved capacity to maintain physiological homeostasis during wide tidal fluctuations of salinity from ~5 to 35 parts per thousand salt. This adaptive plasticity is costly and has limits, so we expected variation among genotypes in their ability to tolerate low salinity (larval survival) and in adult gene expression response to low salinity (Fig. 1). Laura experimentally measured the tolerance of larvae to low salinity after reciprocally conditioning broodstock from high- and low-salinity portions of an estuary (Eierman and Hare 2013). Then, using an oyster transcriptome reference that she assembled de novo (Eierman and Hare 2014), she conducted RNA-seq to measure gene expression differentiation between oyster adults from high- and low-salinity reefs, again tested in a reciprocal transplant tank experiment (Eierman and Hare 2015). Using coding sequence polymorphisms and allele frequencies inferred from the RNAseq data, Laura found a very low average level of genomic differentiation between the two reefs, consistent with high gene flow. However, in the 1% most differentiated loci she also found signatures of directional selection on the low-salinity reef based on nucleotide diversity and allelic correlations among loci. The Hare Lab is continuing to investigate within-generation selection in Delaware Bay oysters in a collaborative NSF-funded project with Daphne Monroe at Rutgers University and Elizabeth North at the Center for Environmental Science, Horn Point Lab, University of Maryland.
Genomics-enabled oyster restoration in New York Harbor Estuary
Eastern oyster (Crassostrea virginica) reefs were abundant in the Hudson/Raritan estuary (HRE; Fig. 2) during and before the colonial period, but were devastated by overfishing and pollution. Oyster reefs are now one of eleven unique habitats prioritized for restoration in the HRE to improve overall environmental quality of the estuary for the benefit of the region. Oyster reefs are valuable habitat used by many species; they help stabilize shorelines and help filter particulates from the water during feeding. If oysters can be successfully recovered to a larger sustainable population size in the HRE, greater resilience is expected for both the environment and its urban inhabitants (local harvest is not the goal).
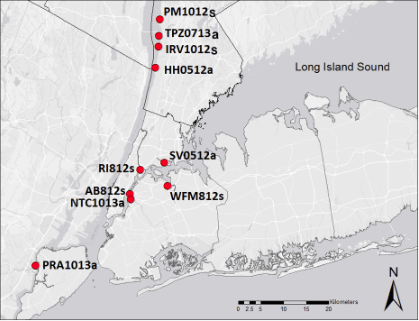
Work in the Hare Lab has been focused on two objectives: (1) testing for genetic population structure among remnant wild oyster populations in the Hudson and East Rivers (Fig. 2) and (2) comparing the fitness-related performance of hatchery-produced oyster cohorts manipulated to have low vs. high genetic diversity. As recent data increasingly showed that a remnant oyster population near the Tappan Zee bridge is reproductively robust (Table 1), we increased efforts to evaluate whether this population can support a population expansion, supplementing hatchery-based restoration. Genomic tests for population structure, funded by New York Sea Grant, were conducted by Yuka Kutsumi for her MS degree. We tested the null hypothesis that juvenile oysters recruiting in HRE were from a genetically homogeneous larval pool, versus an alternative hypothesis that domesticated aquaculture oysters used for pilot-scale restoration projects contributed locally to recruitment. The data support an inference of very high historical gene flow, leading to nearly homogenized allele frequencies across the estuary. However, a genetic discontinuity was found that did not occur near restoration sites as predicted, but instead distinguished East River populations from Hudson River populations with respect to apparent admixture (interbreeding with a genetically different source population). The source of admixture is a question requiring further analyses, but the fact that any differentiation at all occurs between these rivers indicates that larval connectivity is minimal and/or the genetic differences are very recent.
Work on HRE oyster restoration also included a collaborative study funded by the Cornell Atkinson Center for Sustainability (Academic Venture Fund). Efforts to restore oyster beds near urban areas have been slow, in part because public health regulations protecting consumers and the harvest industry are at odds with conservation goals. In addition, hatchery-based population supplementation has been a primary method used to attempt recovery of remnant stocks, but conventional hatchery practices may limit success by reducing genetic diversity of restoration oysters. The project funded by ACSF will improve chances for successful oyster restoration by investigating risk perception among oyster restoration stakeholders (Holley et al. 2018) and testing methods of minimizing hatchery impacts on oyster performance under natural conditions.
Many thanks to Cornell Atkinson for our AVF funding, to the Billion Oyster Project for their cooperation and support, and to our site hosts who keep our experimental oyster cages safe from tampering.
Testing for domestication effects in eastern oysters
There is potential for farmed oysters to contribute to ecosystem services in many of the ways wild oysters do (or used to). However, to the extent that oyster farms are in close proximity to wild oyster populations, and growing oyster strains that were selected for commercially valuable traits, unintended interbreeding between farmed and wild oysters can potentially degrade mean fitness in the wild population. A recent genetic study (Jaris et al. 2019 Conservation Genetics, doi.org/10.1007/s10592-019-01153-9) documented admixture between aquaculture and wild stocks of eastern oysters in a semi-enclosed Rhode Island bay, but the geographic extent of this interaction and its functional consequences are unknown. In theory, as generations of artificial selection increasingly improve traits for production and profit, indirect selection on other traits may change phenotypes (domestication) in ways that are useful for hatchery culture, but maladaptive if the oysters need to complete their life cycle under natural conditions. With collaborators Katie McFarland and Louis Plough at the University of Maryland Center for Environmental Science Horn Point lab, we tested whether starvation tolerance has become weaker in selected oyster strains (constant hatchery propagation) relative to wild oysters. Experiments testing this hypothesis demonstrated robust larval tolerances, and after 10 days with no algal food the survivorship of larvae from wild adults was significantly higher than for larvae from a commercial selected strain. This study was initially funded by a Rapid Response Fund from the Cornell Atkinson Center for Sustainability.
Taking advantage of whole genome sequences and array data for single nucleotide polymorphisms, Hare Lab postdoc Honggang Zhao has been comparing wild source populations with progenitor selected strains to test for genetic differences. Our hypothesis is that many of the largest differences will be due to targeted selection and unique to one or a few independent strains. In addition, if culture conditions are imposing inadvertent selection then we expect some differences to be shared among many of the wild-selected pairs. This work is funded by the Atlantic States Marine Fisheries Commission in collaboration with the eastern oyster breeders consortium.
Diversification at a zoogeographic province boundary
Clustered species range limits in eastern Florida represent a widely recognized boundary between warm-temperate and sub-tropical faunistic provinces. A compression of latitudinal thermoclines along the Florida coast implicates climate as a driving force shaping this community transition, but few studies have directly compared the importance of climate versus hydrographic dispersal barriers or species interactions. The eastern oyster, Crassostrea virginica, is nearly-continuously distributed around Florida, providing an exceptional intraspecific model for distinguishing the relative importance of pre- and postsettlement mechanisms structuring estuarine populations across the province boundary in eastern Florida. A major goal in the Hare lab, previously funded by NSF, is to understand the biotic and abiotic factors that maintain population differentiation in the oyster, and ultimately to test the generality of these influences on other codistributed species.
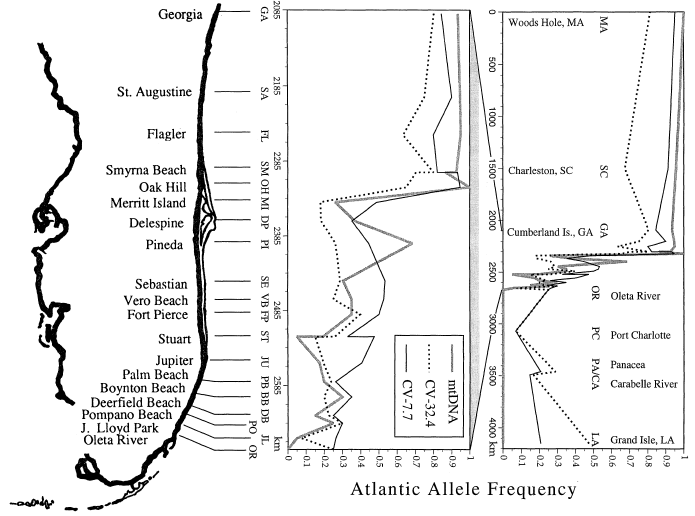
Fig. 5: from Hare and Avise 1996, eastern oyster allele frequency clines for three polymorphisms from Massachusetts to Louisiana (far right), an expanded view from Georgia to Miami (middle), and a map of Florida indicating collection localities along eastern Florida.
Much of the eastern oyster genetic cline in Florida has been stable for the past dozen years (~6 generations; K. Cammen senior thesis). Both dispersal barriers and selection could be maintaining this steep genetic cline. Eastern oysters only occur in the hydrographically semi-closed lagoons behind barrier beaches in eastern Florida, where larval dispersal is predicted to be small-scale, bidirectional and follow a stepping-stone pattern. However, if larvae are flushed out and re-enter lagoons through barrier island inlets, then coastal currents may generate a leap-frog pattern of dispersal. Dispersal directionality could result from larval advection by coastal currents and be a particularly important aspect of cline maintenance, and province boundaries in general, because asymmetrical gene flow can steepen clinal variation and truncate species ranges along a selection gradient (Hare et al. 2005). Multilocus assignment tests on newly-settled oysters were used to measure contemporary gene flow across the step cline and provided no support for recent dispersal across the step cline (Zhang and Hare 2012), consistent with a stable step cline.

The spatial scale of population structure is determined by a balance between dispersal-mediated connectivity, genetic drift, and spatially variable selection. Along the eastern Florida ecotone, we inferred oyster dispersal patterns in combination with the spatial pattern and magnitude of selection as measured by post-settlement cohort analyses and reciprocal transplants. Hatchery-related studies involving oyster spawning and culture were conducted in collaboration with John Scarpa at Harbor Branch Oceanographic Institute. Martha Burford conducted experiments to distinguish three models by which selection may maintain the oyster cline, (1) hybrid unfitness relative to parentals regardless of habitat, (2) habitat-specific hybrid unfitness, and (3) bounded hybrid superiority, with hybrid success constrained to intermediate habitats. She measured fitness-related traits including growth rate, survivorship, rate of reproductive maturation, and infection intensity of a major protozoan parasite, Perkinsus marinus. Martha found that progeny of NxN and SxS crosses showed significant local adaptation (genetic x environment interaction effects) with respect to juvenile survival, rate of reproductive maturation and pre-spawn wet meat weight (Burford et al. 2014). Reproductive barriers were also investigated by Haibin Zhang with in vitro tests of differential cross-fertilization efficiency (Zhang et al. 2010).
Cryptic surf clam subspecies in fisheries and aquaculture

Recently, genetic analyses demonstrated that commercially fished surf clams in Long Island Sound (LIS) are not an ecotype of the Atlantic surf clam, Spisula solidissima solidissima, as previously believed (Hare et al. 2010). Instead, it is a disjunct population of the more southerly surf clam subspecies Spisula solidissima similis, (also known as Raveneli’s surfclam, Spisula raveneli Conrad), a genetically distinct subspecies with no previously known populations occurring north of Cape Hatteras. Initially with funds from USDA Hatch, and continuing with funding from the Mid-Atlantic Fishery Management Council (MAFMC), we are surveying additional habitats in southern New England to document the extent of S.s. similis occurrence, genetically infer the time of origin for these populations, and test for recent hybridization with S.s. solidissima where they co-occur.
With assistance from Dr. Scott Lindell at MBL and aquaculturist Matt Weeks, we acquired a sample of clams from southern Cape Cod and found a mixture of both subspecies with possible hybrids. More recently Mr. Weeks collected surf clams from lagoon-ocean inlets around Long Island, NY (habitat hypothesized to be favored by S.s. similis). Estimation of gene flow between northern and Georgia populations of S.s. similis has been done with mtDNA, microsatellite and nuclear intron markers by Nick Fletcher in the EEB graduate field, showing surprisingly little population differentiation (Fletcher and Hare, in revision). Hannah Hartung is continuing higher resolution genomic analysis of gene flow patterns in both subspecies with MAFMC funding.