This page is continually updated with posts summarizing our latest reports. For background information on the work in our lab, click here: Research Activity
June 23 2022
Testing cats for FIP: updating the ‘Gold Standard’
A new pre-print in Qeios from the Whittaker lab (RNA in-situ hybridization for pathology-based diagnosis of feline infectious peritonitis (FIP): current diagnostics for FIP and comparison to the current gold standard) is now available at https://www.qeios.com/read/NUN8KB. Our research has opened up an opportunity to develop a more sensitive assay for detecting feline coronavirus (FCoV) in tissue samples, opening up an opportunity for new diagnostic technology. Immunohistochemistry (IHC) currently serves as the gold standard for detecting feline coronavirus (FCoV) in an infected cat’s tissue. This study evaluated the efficacy of RNA in situ hybridization (ISH) probes compared to IHC using the monoclonal antibody FIP 3-70. The monoclonal antibody (mAb) is a laboratory-developed protein designed to mimic an immune system’s ability to recognize FIP. For reference, monoclonal antibodies are particular (but hard to develop) laboratory-developed proteins designed to mimic a cat’s immune system’s ability to fight off the virus. The comparison of FIPV 3-70 IHC and RNA ISH using the laboratory probe. The images below were taken of the kidney (large tissue) and the lung (small tissue), under a 4x magnification lens. IHC is shown on the left while RNA ISH is shown on the right (more results can be obtained from the research paper).
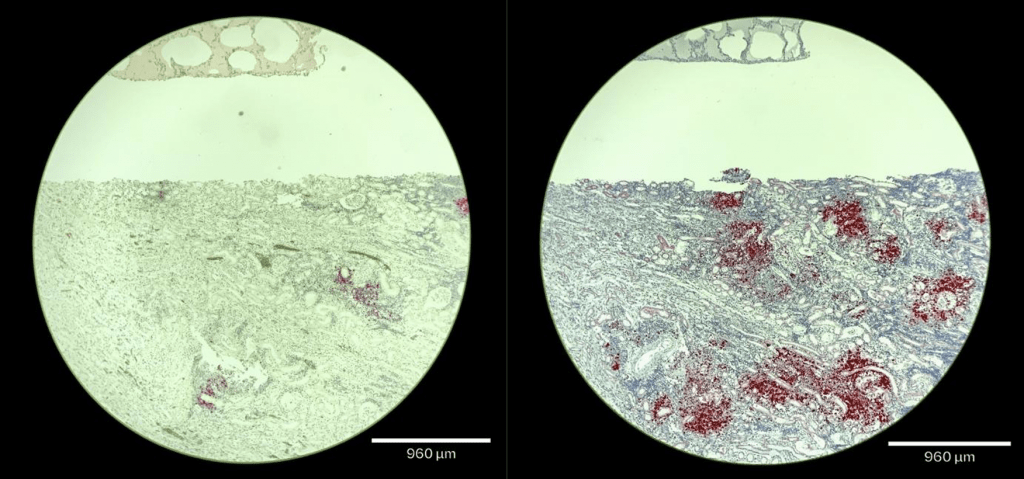
In contrast, RNA probes detect the virus genome and are much more versatile. Comparing the current gold standard of IHC and our new RNA-ISH probe demonstrated that using RNA-ISH markedly improves performance. The improvements from the current gold standard may help researchers and veterinarians better identify FCoV infection in previously undetected cases. More sensitive diagnostic tools could open new ways of understanding FIP. Our improvements from the current gold standard to the sensitivity of FIP diagnosis of the RNA ISH probe need formal validation and have only been tested on post-mortem tissue, but do offer help with clinical research and for veterinarians working closely with FIP-positive felines. The potential benefits of developing new and accurate technology can lead veterinarians and researchers to new treatment options for cats infected with FCoV/FIP.
January 7 2022
An investigation into the evolution of the S1/S2 cleavage motif in murine coronaviruses raises questions about the origins of FCoV
As the New Year begins, the Whittaker Lab is embarking on a new chapter involving the complexities of coronavirus evolution and emergence. For some time now, a focus within our lab was studying how mutations in the S1/S2 domain of the coronaviruses S protein permits cleavage and activation by the enzyme furin, a proprotein convertase found in host cells. In the past, our studies have demonstrated that feline coronaviruses (FCoV); e.g., feline enteric coronavirus (FECV) and feline infectious peritonitis virus (FIPV) are cleaved by furin within the S1/S2 domain at a specific sequence of amino acid residues (SRRSRS) (Licitra et al. 2013). Interestingly, a characteristic of severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) that causes COVID-19 disease has been the equivalent ability of furin to cleave and activate its spike protein (Jaimes et al. 2020). Questions about the evolution of the S1/S2 cleavage site has brought into question the origins of feline coronaviruses, and what animal reservoirs they may have emerged from. Our studies this year have left us with the following question: Bat or Rat?
Rodents often harbor coronaviruses, and in our most recent paper, recently posted in bioRxiv,we discuss the possibility of rodents acting as a putative reservoir for novel coronaviruses, in particular the rodent coronavirus AcCoV-JC34—which comes from the Chevrier’s field mouse in Yunnan Province, China. Interestingly, both SARS-CoV-2 and AcCoV-JC34 are distinct within their subgenera by possessing a unique -RR-RS- predicted furin cleavage site within the S1/S2 domain—likewise FCoV is also distinct in its -RRSRRS- motif. One notable aspect of AcCoV-Jc34 furin cleavage site is that it does not completely align with the S1/S2 motif of most coronavirus spikes, and is instead located in a structurally exposed location above the typical S1/S2 loop. Our analysis suggests that unlike SARS-CoV-2, AcCoV-JC34 may not be cleaved by furin. This contrasts with mouse hepatitis virus (MHV) which has strongly predicted furin cleavage sites. Moving into the New Year, our goal is to shed light on coronavirus evolution and emergence by studying the S1/S2 sites across MHV strains, with an eye to where FCoV may have originated.
Nov 30 2020
SARS-CoV-2 3D structural interactome
In the Whittaker lab, we typically take a highly targeted approach to studying virus biology—with our background knowledge of virology and the insight from the publications of other labs directing a hypothesis-driven approach. However, it is also important to cast a wide net and explore—in what could be described as an untargeted (and arguably unbiased) manner. At the start of the pandemic, we had the pleasure to team up with Haiyuan Yu in the Department of Computational Biology and the Weill Institute for Cell and Molecular Biology at Cornell University. The Yu lab has pioneered approaches to science based on Network Systems Biology, including the development of several webserver tools using machine-learning algorithms (e.g. Interactome INSIDER and mutation3D). In conjunction with the Yu lab, Nicole André in our group is helping deconvolve a new 3D structural interactome of SARS-CoV-2—recently posted to bioRxiv—with one goal being the identification of new functional interactions between the viral proteins and important components of the infected human cells. We hope this will lead to the identification and characterization of novel drug targets, to be used in the ongoing response to COVID-19.
Nov 22 2020
Escaping the cell
To understand how viruses are produced, it is paramount to understand the cells that they infect. In the case of SARS-CoV-2, many labs (including ourselves) are turning to virus-like particles (or pseudoviruses) to limit the need for the high biosafety levels (BSL-3) needed to study the virus itself (as highlighted in JoVE). But it is important to remember that we are using a surrogate virus for these studies and not all viruses exit the cell in the same way. Our recent post of Tiffany Tang’s work highlights some of the issues with the SARS-CoV-2 spike protein, which are of fundamental importance for the many labs studying antibody responses to COVID vaccines.
At the start of the pandemic, we teamed up with cell biologist Nihal Altan-Bonnet at the National Institutes of Health to tackle the issue of exactly how do coronaviruses escape the cell? Dr. Altan-Bonnet’s group, in conjunction with other groups around the world, had been working on a new model for this process using the mouse coronavirus MHV. They discovered that coronaviruses do not use the normal biosynthetic pathway of the cell, but instead harness the power of lysosomes (normally used as a garbage disposer) to exit the cell, and at the same time avoid immune recognition. Marco Straus in the Whittaker lab—using the BSL-3 facilities at Cornell—showed that the pathway also applied to SARS-CoV-2, which sets up fundamentally new models for understanding coronavirus biology and immunology, and provides new targets for anti-viral drug development.
This work was published recently in Cell. More details can also be found in the NIH press release.
Nov 17 2020
Calcium channel blockers show potential as antiviral inhibitors of SARS-CoV-2
There is currently an urgent need for therapeutics to treat COVID-19, and one approach that is being actively investigated in many labs is the re-purposing of FDA-approved drugs that may have addition, or “off-target” activities, and which may be of use for COVID patients. Our previous work on SARS and MERS has led to the realization that there is conserved activity in the coronavirus fusion peptide that relies on binding of calcium ions. Calcium does many things in biology including regulation muscle contraction, and calcium-channel blockers (CCBs) are widely used to treat cardiac problems. To investigate whether CCBs could also inhibit SARS-CoV-2 replication and entry, Marco Straus—in a collaborative project with Susan Daniel— screened a panel of FDA-approved CCBs and found that amlodipine and nifedipine in particular had a good enough profile of efficacy and safety to be considered further.
This study is available in bioRxiv, and as an important next step we have received follow-up FastGrant funding to launch an observational study with Dr. Monica Safford at Weill Cornell Medicine in New York City. We are excited to be doing this type of medical epidemiology to determine whether patients currently taking CCBs may have a benefit when it comes to COVID outcome.
Nov 1 2020
Proteolytic activation of the SARS-CoV-2 spike S1/S2 site: a re-evaluation of furin cleavage
SARS-COV-2 is unique among its closest relatives in that it has a S1/S2 activation site that can be cleaved by furin and furin-like proteases. It shares this feature with several other coronaviruses including MERS-CoV and FCoV serotype I. The Whittaker Lab has teamed up with Dr. Susan Daniel’s in Chemical and Biomedical Engineering to investigate the function of the S1/S2 site in SARS-CoV-2. This study demonstrates that furin cleavage of S1/S2 enhances SARS-CoV-2’s ability to infect human lung cells. While sequencing data from closely related strains suggest that this S1/S2 site is a gain of function, the authors put forward the possibility that SARS-CoV-2 may have evolved from a yet undiscovered parent virus with a more highly active S1/S2 site. This would be similar to research our lab has previously published on loss of S1/S2 furin cleavability in cases of FIP. Tiffany Tang, Javier A. Jaimes, Miya K. Bidon, Marco R. Straus, Susan Daniel and Gary R. Whittaker all contribute to this preliminary report, which is available on bioRxiv.
A detailed summary outlining the methods used in this article – including viral psuedoparticle generation – appears on News-Medical.net.