Comparison of Sanger sequencing vs. Next Generation Sequencing
Contributed by Faith Meza, edited by Beth Licitra and Ximena Andrea Olarte Castillo
No matter in which field you work, you have probably heard of Sanger sequencing and next generation sequencing (NGS) or have experience using them. With new sequencing technology revolutionizing science and medicine, it can be hard to keep up with the best DNA and RNA sequencing methods available. In this post, we will be covering the similarities and differences between Sanger and NGS sequencing.
The Sanger sequencing method was first developed in the early 1990s by the English biochemist, Frank Sanger. It relies on capillary electrophoresis technology in which each nucleotide is assigned a unique fluorescent marker. Sanger sequencing is very accurate, but it can only sequence a single strand of DNA at a time. On the other hand, NGS is an improvement on Sanger sequencing in that it allows for the sequencing of many DNA or RNA strands simultaneously. Currently, several NGS platforms exists (i.e Oxford Nanopore, Illumina, PacBio) each of which allow the users to sequence with a different resolution, accuracy and sample turn out. In the field of virology, NGS has been widely used to identify and characterize viruses in a sample without prior knowledge of the virus sequence (i.e. non-specific or PCR-free sequencing). This has resulted in a recent increase of viral discovery in samples from humans, animals, plants, deep sea sediments and more. NGS technologies can also be used in targeted methods, like the sequencing of PCR products, in which the high throughput results of the sequencing can reveal the complex composition of a viral population in a sample.
Comparison of Sanger Sequencing and Next Generation Sequencing
Sanger Sequencing |
Next generation sequencing |
|
Benefits |
● Can run a sample in 24 hours
● Cost Effective ● 1-20 targets ● 99.9% accurate |
● Higher sequencing ability
● Higher discovery power ● Higher mutation resolution (ability to catch the mutations of DNA strains) ● Higher sample turn out ● Can run millions of DNA fragments |
Challenges |
● Low sensitivity
● Low discovery power ● Not as time efficient ● Unable to meet higher output demands |
● High initial cost
● Lower accuracy rate ● Error rates range between 0.1 and 1% |
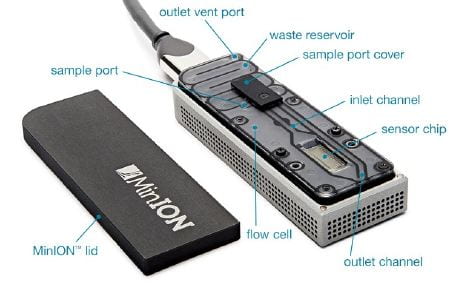
Our work in the Whittaker lab is shifting from Sanger sequencing to NGS to allow us to detect subtle variations in viral populations within an affected cat. NGS technology has also allowed us to analyze larger portions of the viral genome to study a wider variety of genes and genetic changes that are suspected to be associated with the transition to FIP. We are currently using the Oxford Nanopore sequencing technology, which has a convenient portable sequencing machine (the MinION) in which it is possible to see sequencing results in real time. We have used this technology to sequence specific regions of interest in the Spike protein of feline coronavirus to develop a risk assessment test to aid in the diagnosis of FIP.