 |
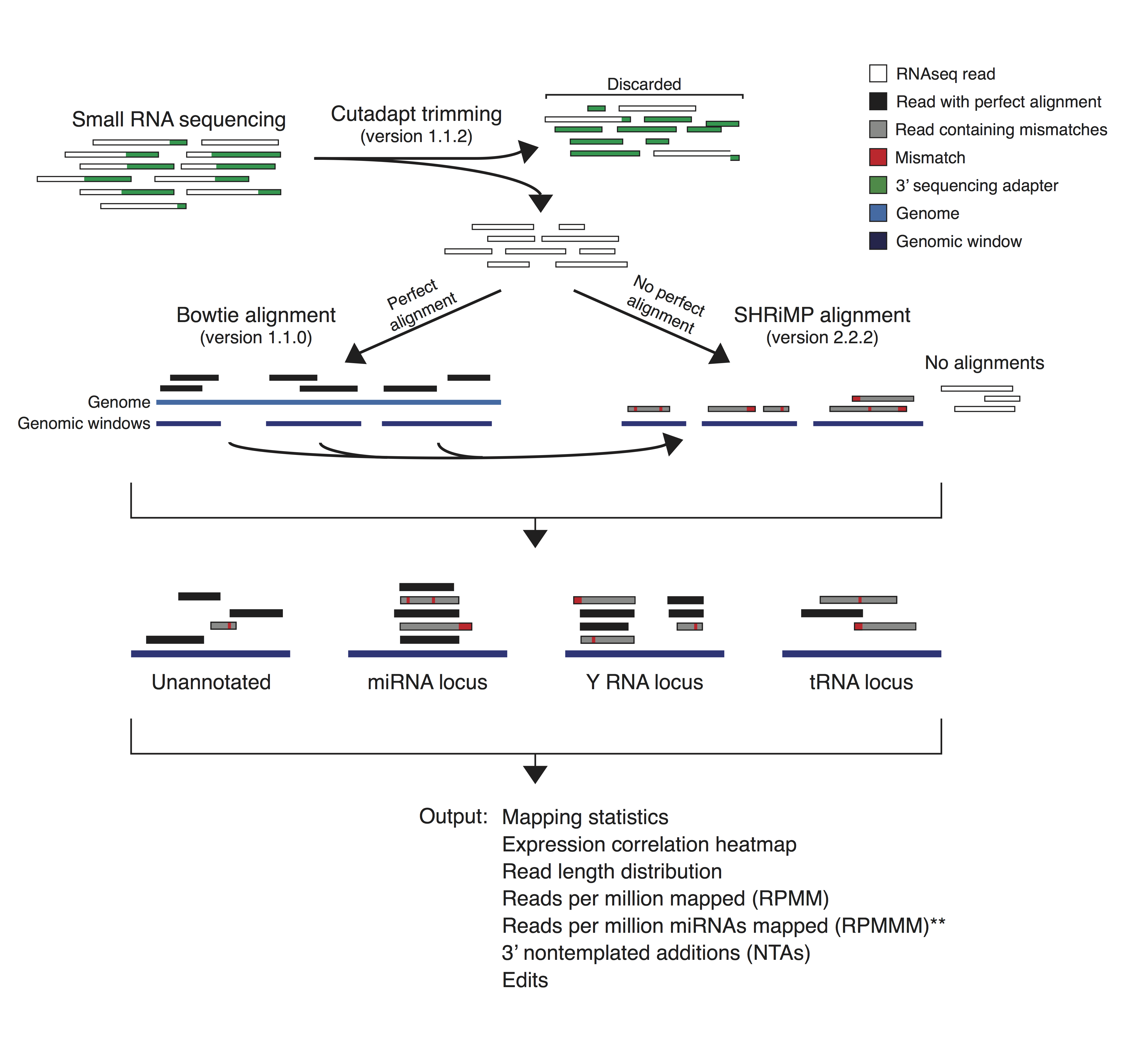
miRquant 2.0 Description: This program takes as input small RNA-seq data (human, mouse, and rat) and provides a detailed annotation and quantification of microRNAs and their isomiRs. The comprehensive output includes mapping statistics, read length distributions, relative abundance of different small RNA types, normalized expression levels of miRNAs, and quantification of 3′ non-templates nucleotide addition and RNA editing events. Code and tutorial: https://github.com/Sethupathy-Lab/miRquant Citation: Kanke et al., 2016, Journal of Integrative Bioinformatics |
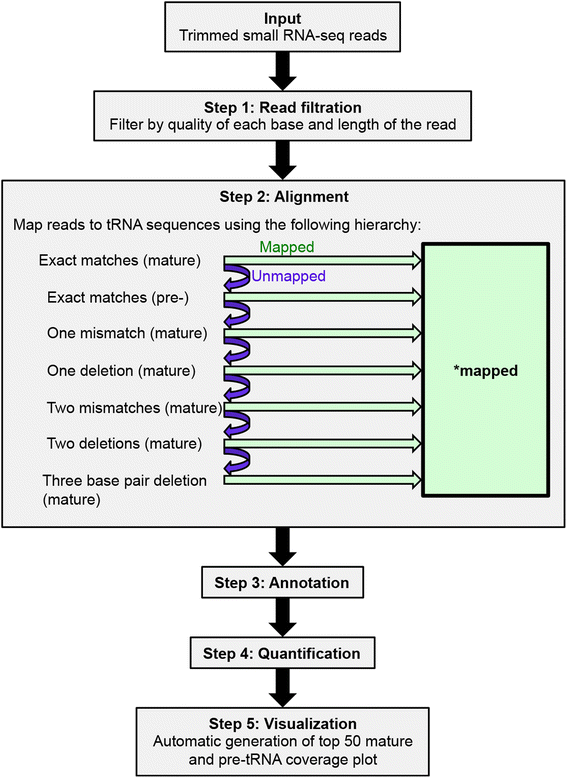 |
tDRmapper Description: A tool for mapping, naming, and quantifying tRNA-derived RNAs (tDRs). tDRmapper was designed specifically for human small RNA-seq data (single-end, 50x) generated on the Illumina HiSeq platform using cDNA libraries that were prepared using the Illumina TruSeq protocol. It not only provides a standardized nomenclature and quantification scheme, but also includes graphical visualization that facilitates the discovery of novel transfer RNAs and tRNA-derived RNA biology. Code: https://github.com/sararselitsky/tDRmapper Citation: Selitsky and Sethupathy, 2016, BMC Bioinformatics |
