Metabolic Networks
Bioreactors are systems that use an organism to perform some industrial function; typically, they involve a tank full of Escherichia coli (not the infectious kind, but the harmless lab strain) that are engineered to produce some useful chemical or protein. However, early on in bioengineering, bioreactors ran into a huge problem, particularly when they fed the bacteria with glucose (which they wanted to do because it made the system well-defined, and thus predictable– and glucose is cheap and very efficient): a buildup of acetate caused the reactor to become rather acidic and start killing off the bacteria.
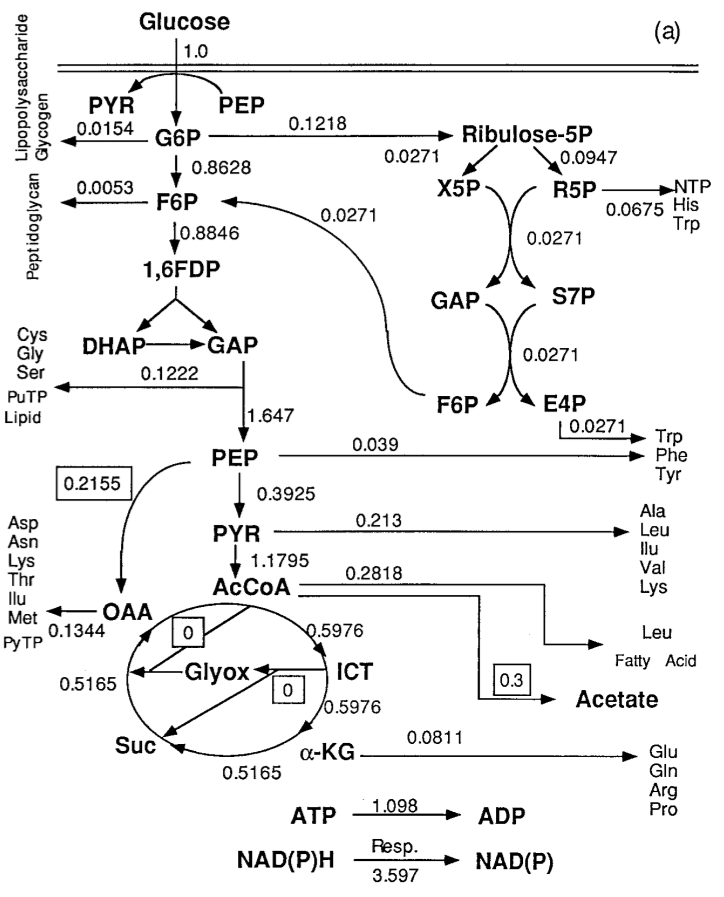
In order to understand why acetate built up, researchers built a partial model, or a a metabolic network, of E. coli‘s metabolism, specifically of glycolysis and the citric acid cycle. In this network, nodes represented a metabolite, such as glucose, glucose-phosphate, or acetate, and edges represented the reactions that converted them, as catalyzed by specific enzymes. The edges were labeled with experimentally determined metabolic flux, which is essentially the rate at which metabolites flow from one node to another. Here’s what it looked like:
What they learned from this is that there were two key ways they might be able to reduce acetate production: increasing the rate of the PEP (phosphoenolpyruvate) to OAA (oxaloacetate) reaction, or increasing the Glyox (glyoxylate bypass) edges. In a manner similar to what we’ll be seeing later with cascading behavior in networks, the researchers modeled this; their model predicted that acetate production would decrease, and metabolites used for proteins would increase. When they tested this in actual engineered strains, they found that in the increased PEP->OAA strain, not only did it decrease acetate production, but it increased the flux towards metabolites that could be used to make proteins, increasing the efficiency of conversion from glucose to protein. However, the increased Glyox strain, although acetate production still decreased, efficiency of glucose conversion to protein was lower than in normal E. coli. These results highlight both the strength and the limitations of network analyses for metabolism; although some useful predictions can be obtained, an incomplete metabolic network model is likely to leave out interactions that have real-life effects on the cells.
~rezecib