 A new technology called rhAmpSeq™ is allowing grape
A new technology called rhAmpSeq™ is allowing grape
geneticists and breeders to rapidly find and validate 2000 markers in diverse grape varieties and species. These ‘core genome markers’ provide a new set of mileposts that were used during a pilot study by the USDA-funded VitisGen2 project to map traits in six unrelated ‘mapping populations’ representing the diversity of U.S. grape breeding programs, including numerous Vitis species.
The rhAmpSeq technology, developed by Integrated DNA Technologies (IDT) and commercialized in March 2019, allows researchers to mix and amplify DNA from up to 4,000 individual samples and simultaneously sequence thousands of different markers of each sample. This dramatically reduces the cost of DNA sequencing across the 19 grape chromosomes – and offers a detailed map of the entire genome for each of the 4000 individuals in one batch.
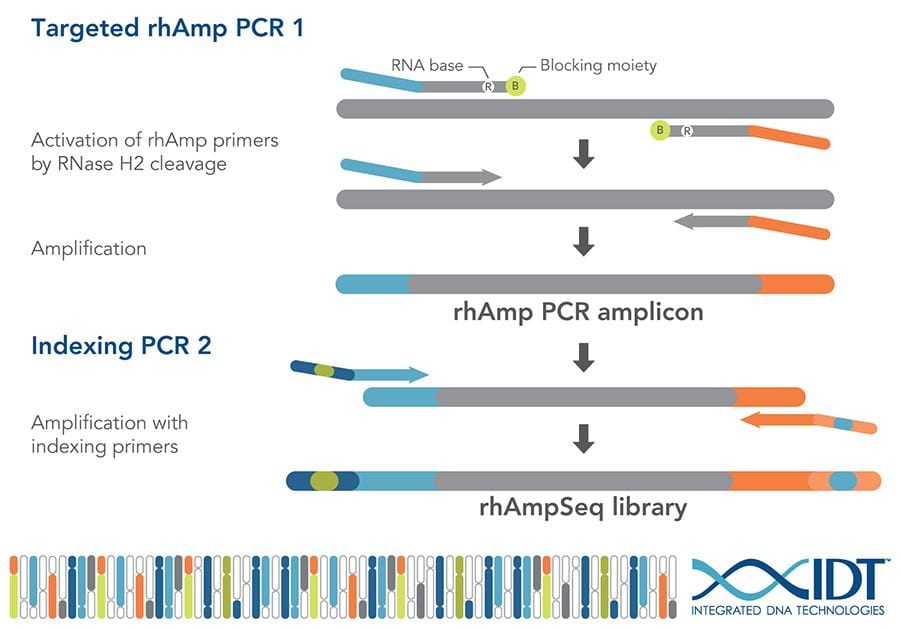
The rhAmpSeq technology uses normal DNA primers followed by an RNA base and a PCR blocker in PCR 1. After these primers bind specifically to the target DNA, the RNA and blocker are removed and PCR amplification continues. This improved specificity enables thousands of markers to be amplified simultaneously for a given sample. Then ‘barcoding’ primers are used in PCR 2 to label each sample uniquely prior to pooling all samples for sequencing. This allows amplification and sequencing of 2000 DNA markers in up to 4000 samples.
The VitisGen2 project team, a multi-institutional research collaboration funded by the USDA-National Institute of Food and Agriculture Specialty Crops Research Initiative, played a key role in adapting the rhAmpSeq process to plant breeding platforms. The VitisGen2 approach provided two key improvements to the AmpSeq technology developed during the first VitisGen project: 1) improved marker design after sequencing 7 new genomes, which drastically improved marker transferability across diverse breeding lines, and 2) the rhAmpSeq marker chemistry itself, which improves the specificity of AmpSeq, enabling more markers to be multiplexed in a single PCR (from 400 markers with AmpSeq to 2000 markers with rhAmpSeq). Unlike single nucleotide markers that provide binary information (reference allele or alternative allele), the larger 250 base pair sequences employed by VitisGen2’s rhAmpSeq strategy generates high information content per marker.
Professor Bruce Reisch commented, “As early testers of the rhAmpSeq system from IDT, we were really pleased to see how we were able to rapidly accelerate the VitisGen2 program by using a nearly 2,000 marker rhAmpSeq panel to analyze 19 Vitis linkage groups. The core marker set was useful across six unrelated populations representing the diversity of the genus, and the workflow was very easy to use and allowed high-throughput processing.”
“We sought to develop an amplicon sequencing technology that addressed the unmet needs of researchers—increased specificity of PCR amplification, while minimizing primer dimer formation,” said Yongming Sun, IDT’s bioinformatics senior staff scientist. “Through collaboration with our diverse group of beta testers, including Cornell University’s College of Agriculture and Life Sciences, we developed the rhAmpSeq Amplicon Sequencing System, which allows researchers to achieve a high degree of multiplexing capability with thousands of assays in a single tube.”
Although the VitisGen2 project team helped pioneer this use of the technology, other scientists and plant breeders see the technology as being broadly applicable to a wide variety of crops.
For more information, you can read the March 6 news release from IDT: https://idtb.io/38tvsg
The VitisGen2 project is supported by the USDA-NIFA Specialty Crop Research Initiative (Award No. 2017- 51181-26829).

I didn’t know that the technology mentioned in this article could allow people to mix and amplify DNA. My husband has been wanting to learn more about DNA sequencing ever since hearing about mutated viruses. I think he should look into someone with experience in next-generation DNA sequencing that could answer some of his questions.
This is a research tool, requiring specialized equipment, and designed to answer research questions about plants. Thanks for your interest.
Any kind of computer requirements (or limitations) people should be aware of so they (or a computer savvy student ) could take off running after learning more about this?
Thanks!
Thanks for your interest. Avi Karn is our bioinformatics specialist, and he works with the rhAmpSeq data and our ‘mapping populations’ to map the plant phenotypes and associate them with DNA markers. The software is specialised – and associated with the data acquisition. Avi would have more information about it.
‘