Our work focuses on the evolution of butterfly wing patterns. We are specifically interested in how novel patterns arise in nature and how they diversify under natural selection. Variation is the raw material of evolution and is the most fundamental determinant of morphological diversity. Little is known, however, about the developmental genetic forces that shape the range of color pattern variation appearing in natural populations.
We approach our study system at multiple levels of organization, ranging from the developmental genetic basis of color pattern formation to the adaptive roles color patterns play in nature. One of the major benefits of working on butterflies is that many wing patterns have clear field-tested adaptive value, especially in cases of mimicry, camouflage, and mate preference. Furthermore, many butterfly species show significant geographic variation that allows mapping and positional cloning of genes underlying natural pattern variation. These advantages make it possible to study the developmental origins of variation, along with the effects of natural selection on that variation, in a way that is difficult in many model organisms.
Developmental genetics of mimicry
Heliconius butterflies are a textbook model system in evolutionary biology and have provided many key insights into the evolutionary genetics of mimicry and color pattern evolution. Genetic mapping has revealed that only a few genomic regions underlie most of the natural variation resulting in the hundreds of distinct wing patterns seen in the genus. These loci behave as genomic hotspots of evolution because they drive adaptive variation within and between species with both convergent and highly divergent wing patterns, thus demonstrating that some genes are unusually prone to facilitate rapid evolutionary change. We are working to characterize the genes underlying mimicry, and to understand how they evolve functionally. Two genes of particular interest to our lab are optix and WntA.
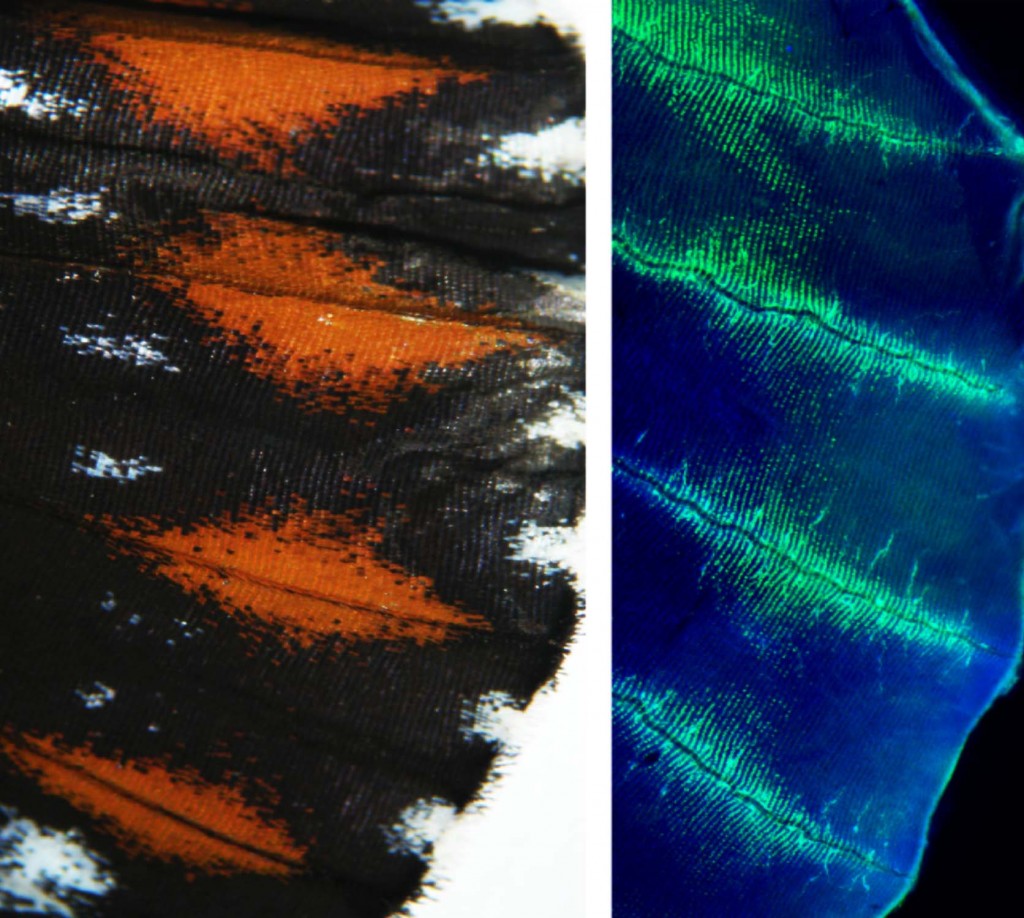
The role of optix in the evolution of complex red color patterns
In 2011 we reported the discovery of the first Heliconius color pattern gene. This gene, optix, is a homeobox transcription factor that controls within- and between-species variation of complex red wing patterns. This work is of general interest because it shows how cis-regulatory evolution of a single gene can drive the adaptation of complex color patterns. It also shows that the same gene can repeatedly underlie pattern evolution across highly divergent lineages.
Black patterns evolve through regulatory variation of a morphogen gene
It has long been speculated that spatial changes in cell-signaling gradients should underlie pattern evolution, however there has been little direct evidence for this. For the first time we demonstrate that positional variation in an inductive signal is responsible for pattern evolution within and between species. Specifically, we found that the morphogen WntA induces complex black color patterns in Heliconius, and that distinct cis-regulatory alleles of WntAare responsible for mimicry-related patterns in local populations. This work shows that a morphogen gene itself can be a source of heritable pattern variation in nature.
The wing pattern ground plan
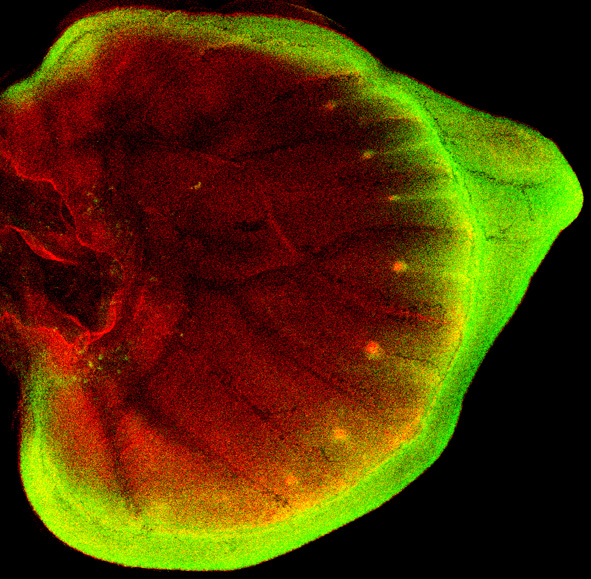
Butterfly wing patterns evolve largely through the gain, loss, and alteration of elements derived from a conserved ground plan of spots and stripes. It is possible to trace ground plan changes to specific butterfly lineages and to investigate the developmental basis of those changes. In this context we have shown that the evolution of Notch, Distal-less, Spalt, and Engrailed regulation is associated with the diversification of eyespot-related ground plan elements. Additionally we have shown that wingless is implicated in stripe patterning, and that a Lepidoptera-specific duplicate of aristaless prefigures one conserved stripe element. This is one of the first examples of how the origin of a discrete pattern element can be associated with duplication and regulatory neofunctionalization of a regulatory gene.
Evolutionary origin and adaptive function of wing pigmentation

Our lab has a long term interest in the origin and function of butterfly coloration. We have shown that the origin of red and orange wing pigments in most butterflies can be attributed to regulatory co-option of ommochrome eye pigment genes such as cinnabar and vermilion. We are also working to describe new pigments and pigment-related genes that are expressed in butterfly wings. Recent collaborative work with Adriana Briscoe and others has revealed co-evolution of wing pigmentation and opsin genes, illuminating a fascinating example of the co-evolution of a visual signal and receptor.
Regulatory architecture of color pattern evolution
Despite an intense interest in the evolution of animal morphology, very little is known about how complex patterns originate and evolve in nature. Indeed, most examples of genes driving phenotypic evolution are case studies of simple changes such as trait loss or reduction that are caused by a compromise of gene function. In this light, our recent identification of two genes controlling adaptation of complex butterfly wing patterns raises a new set of questions about pattern evolution. For both genes – optix and WntA – the functional variation is cis-regulatory, therefore much of our current research is focused on the issue of how the evolution of cis-regulatory elements can drive the rapid evolution of highly complex expression patterns.
Wing patterns, mimicry, and social behavior in Heliconius butterflies
Heliconius wing patterns play a role in predator deterrence and mate choice, however it is not known precisely what wing pattern elements avian predators respond to – e.g. is it color, contrast, shape, size, or something else? This question is relevant because we know specific genes control particular characteristics of wing patterns and therefore we may be able to determine which predator behaviors are driving selection at various different loci. To explore this we have been working with Adriana Briscoe to develop optically accurate butterfly wing models to use in fieldwork. In our first study using these models we determined that Heliconius communal roosts strongly deter predation, thus suggesting that pattern frequency is an important parameter in defining the fitness benefits of wing patterns. Furthermore, we were surprised to find that there is an optimal roost size for maximizing predator deterrence that closely corresponds with observed roost sizes in nature. We are now using these tools to dissect the fundamental elements of butterfly aposematism in nature by measuring predation rates on models with altered color patterns.
Evolution of color pattern plasticity

In many species individuals that are genetically similar can develop into one of several distinct morphs in response to environmental cues. This phenomenon, commonly known as polyphenism, is significant because it can allow rapid adaptation of organisms to their environments without relying on genetic changes between generations. This topic is of fundamental interest, however major questions remained unanswered, especially regarding how polyphenisms evolve. While much work has been done on the mechanisms and fitness advantages of polyphenism, less has been done addressing how polyphenisms can vary between populations to facilitate adaptation to local conditions. We are addressing these questions using seasonal color variation in buckeye butterflies. We have shown that there is dramatic variation in developmental plasticity between populations, with west coast buckeyes being robust to environmental variation while east coast buckeyes show a strong response to cool temperatures and long day lengths. We are now analyzing a large RNA-seq dataset to assess what kind of gene expression differences underlie the different polyphenic phenotypes. We are also working to make crosses between populations to map the gene(s) controlling regional variation in polyphenism.